Project flow¶
LaminDB allows tracking data lineage on the entire project level.
Here, we walk through exemplified app uploads, pipelines & notebooks following Schmidt et al., 2022.
A CRISPR screen reading out a phenotypic endpoint on T cells is paired with scRNA-seq to generate insights into IFN-γ production.
These insights get linked back to the original data through the steps taken in the project to provide context for interpretation & future decision making.

More specifically: Why should I care about data flow?
Data flow tracks data sources & transformations to trace biological insights, verify experimental outcomes, meet regulatory standards, increase the robustness of research and optimize the feedback loop of team-wide learning iterations.
While tracking data flow is easier when it’s governed by deterministic pipelines, it becomes hard when it’s governed by interactive human-driven analyses.
LaminDB interfaces workflow mangers for the former and embraces the latter.
# !pip install 'lamindb[jupyter,bionty,aws]'
!lamin init --storage ./mydata
Show code cell output
💡 connected lamindb: testuser1/mydata
Import lamindb:
import lamindb as ln
from IPython.display import Image, display
💡 connected lamindb: testuser1/mydata
Steps¶
In the following, we walk through exemplified steps covering different types of transforms (Transform).
Note
The full notebooks are in this repository.
App upload of phenotypic data  ¶
¶
Register data through app upload from wetlab by testuser1:
# This function mimics the upload of artifacts via the UI
# In reality, you simply drag and drop files into the UI
def mock_upload_crispra_result_app():
ln.setup.login("testuser1")
transform = ln.Transform(name="Upload GWS CRISPRa result", type="upload")
ln.track(transform=transform)
output_path = ln.core.datasets.schmidt22_crispra_gws_IFNG(ln.settings.storage.root)
output_file = ln.Artifact(
output_path, description="Raw data of schmidt22 crispra GWS"
)
output_file.save()
mock_upload_crispra_result_app()
Show code cell output
💡 saved: Transform(uid='uUkmDk4oAZThcxpd', name='Upload GWS CRISPRa result', type='upload', created_by_id=1, updated_at='2024-08-05 13:27:39 UTC')
💡 saved: Run(uid='MyJQISjBlEDBL0Wpm2Ss', transform_id=1, created_by_id=1)
Hit identification in notebook  ¶
¶
Access, transform & register data in drylab by testuser2 in notebook hit-identification.
Show code cell content
# the following mimics the integrated analysis notebook
# In reality, you would execute inside the notebook
import nbproject_test
from pathlib import Path
cwd = Path.cwd()
nbproject_test.execute_notebooks(cwd / "project-flow-scripts/hit-identification.ipynb", write=True)
Executing notebooks in /home/runner/work/lamin-usecases/lamin-usecases/docs/project-flow-scripts/hit-identification.ipynb
Scheduled: ['hit-identification']
hit-identification
✓ (3.922s)
Total time: 3.924s
Inspect data flow:
artifact = ln.Artifact.filter(description="hits from schmidt22 crispra GWS").one()
artifact.view_lineage()
Sequencer upload  ¶
¶
Upload files from sequencer via script chromium_10x_upload.py:
!python project-flow-scripts/chromium_10x_upload.py
Show code cell output
💡 connected lamindb: testuser1/mydata
💡 saved: Transform(uid='qCJPkOuZAi9q5zKv', version='1', name='chromium_10x_upload.py', key='chromium_10x_upload.py', type='script', created_by_id=1, updated_at='2024-08-05 13:27:45 UTC')
💡 saved: Run(uid='M1YUHtoMegy8j1ovlcze', transform_id=3, created_by_id=1)
scRNA-seq bioinformatics pipeline  ¶
¶
Process uploaded files using a script or workflow manager: Pipelines – workflow managers and obtain 3 output files in a directory filtered_feature_bc_matrix/:
!python project-flow-scripts/cellranger.py
Show code cell output
💡 connected lamindb: testuser1/mydata
💡 saved: Transform(uid='VABfOZzDfL3JFBf2', version='7.2.0', name='Cell Ranger', type='pipeline', reference='https://www.10xgenomics.com/support/software/cell-ranger/7.2', created_by_id=2, updated_at='2024-08-05 13:27:47 UTC')
💡 saved: Run(uid='JnrWwqx9rnDqRP7Q2KT5', transform_id=4, created_by_id=2)
❗ this creates one artifact per file in the directory - consider ln.Artifact(dir_path) to get one artifact for the entire directory
!python project-flow-scripts/postprocess_cellranger.py
Show code cell output
💡 connected lamindb: testuser1/mydata
💡 saved: Transform(uid='YqmbO6oMXjRj65cN', version='2', name='postprocess_cellranger.py', key='postprocess_cellranger.py', type='script', created_by_id=2, updated_at='2024-08-05 13:27:49 UTC')
💡 saved: Run(uid='444H99fl2Aj6URgoDD5P', transform_id=5, created_by_id=2)
Inspect data flow:
output_file = ln.Artifact.filter(description="perturbseq counts").one()
output_file.view_lineage()
Integrate scRNA-seq & phenotypic data  ¶
¶
Integrate data in notebook integrated-analysis.
Show code cell content
# the following mimics the integrated analysis notebook
# In reality, you would execute inside the notebook
nbproject_test.execute_notebooks(cwd / "project-flow-scripts/integrated-analysis.ipynb", write=True)
Executing notebooks in /home/runner/work/lamin-usecases/lamin-usecases/docs/project-flow-scripts/integrated-analysis.ipynb
Scheduled: ['integrated-analysis']
integrated-analysis
✓ (4.185s)
Total time: 4.186s
Review results¶
Let’s load one of the plots:
# track the current notebook as transform
ln.settings.transform.stem_uid = "1LCd8kco9lZU"
ln.settings.transform.version = "0"
ln.track()
💡 notebook imports: ipython==8.26.0 lamindb==0.75.0 nbproject_test==0.5.1
💡 saved: Transform(uid='1LCd8kco9lZU6K79', version='0', name='Project flow', key='project-flow', type='notebook', created_by_id=1, updated_at='2024-08-05 13:27:55 UTC')
💡 saved: Run(uid='65p6C3tNUVbb2hn2BKFu', transform_id=7, created_by_id=1)
Run(uid='65p6C3tNUVbb2hn2BKFu', started_at='2024-08-05 13:27:55 UTC', is_consecutive=True, transform_id=7, created_by_id=1)
artifact = ln.Artifact.filter(key__contains="figures/matrixplot").one()
artifact.cache()
Show code cell output
PosixUPath('/home/runner/work/lamin-usecases/lamin-usecases/docs/mydata/.lamindb/bHNw9mwCIrWb8My59UQN.png')
display(Image(filename=artifact.path))
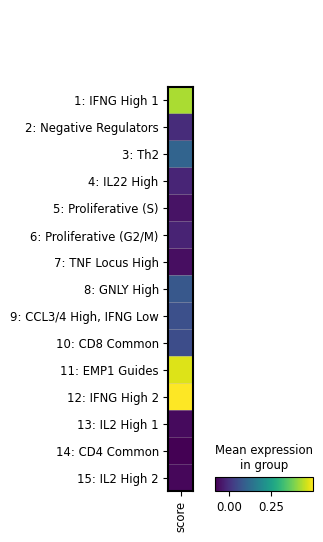
We see that the image artifact is tracked as an input of the current notebook. The input is highlighted, the notebook follows at the bottom:
artifact.view_lineage()
Alternatively, we can also look at the sequence of transforms:
transform = ln.Transform.search("Project flow").first()
transform.predecessors.df()
| uid | version | name | key | description | type | source_code | hash | reference | reference_type | _source_code_artifact_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 6 | lB3IyPLQSmvt5zKv | 1 | Perform single cell analysis, integrate with C... | integrated-analysis | None | notebook | None | None | None | None | None | 2 | 2024-08-05 13:27:53.636178+00:00 |
transform.view_lineage()
Understand runs¶
We tracked pipeline and notebook runs through run_context, which stores a Transform and a Run record as a global context.
Artifact objects are the inputs and outputs of runs.
What if I don’t want a global context?
Sometimes, we don’t want to create a global run context but manually pass a run when creating an artifact:
run = ln.Run(transform=transform)
ln.Artifact(filepath, run=run)
When does an artifact appear as a run input?
When accessing an artifact via cache(), load() or open(), two things happen:
The current run gets added to
artifact.input_ofThe transform of that artifact gets added as a parent of the current transform
You can then switch off auto-tracking of run inputs if you set ln.settings.track_run_inputs = False: Can I disable tracking run inputs?
You can also track run inputs on a case by case basis via is_run_input=True, e.g., here:
artifact.load(is_run_input=True)
Query by provenance¶
We can query or search for the notebook that created the artifact:
transform = ln.Transform.search("GWS CRIPSRa analysis").first()
And then find all the artifacts created by that notebook:
ln.Artifact.filter(transform=transform).df()
| uid | version | description | key | suffix | type | _accessor | size | hash | _hash_type | n_objects | n_observations | visibility | _key_is_virtual | storage_id | transform_id | run_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||
| 2 | gLnzC5usf15rLUVLgz2b | None | hits from schmidt22 crispra GWS | None | .parquet | dataset | DataFrame | 18368 | ky0pLx8o9oFJnn0niPUHog | md5 | None | None | 1 | True | 1 | 2 | 2 | 2 | 2024-08-05 13:27:43.193138+00:00 |
Which transform ingested a given artifact?
artifact = ln.Artifact.filter().first()
artifact.transform
Transform(uid='uUkmDk4oAZThcxpd', name='Upload GWS CRISPRa result', type='upload', created_by_id=1, updated_at='2024-08-05 13:27:39 UTC')
And which user?
artifact.created_by
User(uid='DzTjkKse', handle='testuser1', name='Test User1', updated_at='2024-08-05 13:27:45 UTC')
Which transforms were created by a given user?
users = ln.User.lookup()
ln.Transform.filter(created_by=users.testuser1).df()
| uid | version | name | key | description | type | source_code | hash | reference | reference_type | _source_code_artifact_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 1 | uUkmDk4oAZThcxpd | None | Upload GWS CRISPRa result | None | None | upload | None | None | None | None | NaN | 1 | 2024-08-05 13:27:39.020048+00:00 |
| 3 | qCJPkOuZAi9q5zKv | 1 | chromium_10x_upload.py | chromium_10x_upload.py | None | script | None | None | None | None | 5.0 | 1 | 2024-08-05 13:27:45.599434+00:00 |
| 7 | 1LCd8kco9lZU6K79 | 0 | Project flow | project-flow | None | notebook | None | None | None | None | NaN | 1 | 2024-08-05 13:27:55.261966+00:00 |
Which notebooks were created by a given user?
ln.Transform.filter(created_by=users.testuser1, type="notebook").df()
| uid | version | name | key | description | type | source_code | hash | reference | reference_type | _source_code_artifact_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 7 | 1LCd8kco9lZU6K79 | 0 | Project flow | project-flow | None | notebook | None | None | None | None | None | 1 | 2024-08-05 13:27:55.261966+00:00 |
We can also view all recent additions to the entire database:
ln.view()
Show code cell output
Artifact
| uid | version | description | key | suffix | type | _accessor | size | hash | _hash_type | n_objects | n_observations | visibility | _key_is_virtual | storage_id | transform_id | run_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||
| 12 | bHNw9mwCIrWb8My59UQN | None | None | figures/matrixplot_fig2_score-wgs-hits-per-clu... | .png | None | None | 28814 | vKSAeP8dZnBAPkdOYa_kRQ | md5 | None | None | 1 | True | 1 | 6 | 6 | 2 | 2024-08-05 13:27:54.473110+00:00 |
| 11 | RJNnQ23OK9kG6wBGLYAC | None | None | figures/umap_fig1_score-wgs-hits.png | .png | None | None | 118999 | IlWQvuhi-VqBf1nCqWnYXQ | md5 | None | None | 1 | True | 1 | 6 | 6 | 2 | 2024-08-05 13:27:54.283715+00:00 |
| 10 | wiwLZKKJVRIBOkouHiYc | None | perturbseq counts | schmidt22_perturbseq.h5ad | .h5ad | None | AnnData | 20659936 | la7EvqEUMDlug9-rpw-udA | md5 | None | None | 1 | False | 1 | 5 | 5 | 2 | 2024-08-05 13:27:50.448002+00:00 |
| 9 | m9a4XNWIBHL9PSous5gn | None | None | perturbseq/filtered_feature_bc_matrix/matrix.m... | .mtx.gz | None | None | 6 | rMd6znmJIBUQQqzq9GpsSg | md5 | None | None | 1 | False | 1 | 4 | 4 | 2 | 2024-08-05 13:27:47.944693+00:00 |
| 8 | GG8apR81yeBE1XionLLQ | None | None | perturbseq/filtered_feature_bc_matrix/barcodes... | .tsv.gz | None | None | 6 | fEi3nrFvLf0Pfm3fXYN7Ag | md5 | None | None | 1 | False | 1 | 4 | 4 | 2 | 2024-08-05 13:27:47.944099+00:00 |
| 7 | 1pdhaiNC1mYg06lM9O24 | None | None | perturbseq/filtered_feature_bc_matrix/features... | .tsv.gz | None | None | 6 | mFJD9xzjZiqfprVRXP82YQ | md5 | None | None | 1 | False | 1 | 4 | 4 | 2 | 2024-08-05 13:27:47.943210+00:00 |
| 4 | tVB61tpZ64kZ7fsaZavQ | None | None | fastq/perturbseq_R2_001.fastq.gz | .fastq.gz | None | None | 6 | _FYwYQexjj1IdytzDh4r3A | md5 | None | None | 1 | False | 1 | 3 | 3 | 1 | 2024-08-05 13:27:45.583004+00:00 |
Run
| uid | started_at | finished_at | is_consecutive | reference | reference_type | transform_id | report_id | environment_id | parent_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||
| 1 | MyJQISjBlEDBL0Wpm2Ss | 2024-08-05 13:27:39.023583+00:00 | NaT | True | None | None | 1 | None | NaN | None | 1 |
| 2 | rBly8PYutQjczzmLdEuw | 2024-08-05 13:27:42.780636+00:00 | NaT | True | None | None | 2 | None | NaN | None | 2 |
| 3 | M1YUHtoMegy8j1ovlcze | 2024-08-05 13:27:45.222022+00:00 | 2024-08-05 13:27:45.597764+00:00 | True | None | None | 3 | None | 6.0 | None | 1 |
| 4 | JnrWwqx9rnDqRP7Q2KT5 | 2024-08-05 13:27:47.571179+00:00 | NaT | None | None | None | 4 | None | NaN | None | 2 |
| 5 | 444H99fl2Aj6URgoDD5P | 2024-08-05 13:27:49.433758+00:00 | NaT | None | None | None | 5 | None | NaN | None | 2 |
| 6 | vDLvDu4nSAVn9dAIsi9v | 2024-08-05 13:27:53.641972+00:00 | NaT | True | None | None | 6 | None | NaN | None | 2 |
| 7 | 65p6C3tNUVbb2hn2BKFu | 2024-08-05 13:27:55.267325+00:00 | NaT | True | None | None | 7 | None | NaN | None | 1 |
Storage
| uid | root | description | type | region | instance_uid | run_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||
| 1 | Ag1p7yzENP47 | /home/runner/work/lamin-usecases/lamin-usecase... | None | local | None | 54ZGqgkROOFf | None | 1 | 2024-08-05 13:27:37.306737+00:00 |
Transform
| uid | version | name | key | description | type | source_code | hash | reference | reference_type | _source_code_artifact_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 7 | 1LCd8kco9lZU6K79 | 0 | Project flow | project-flow | None | notebook | None | None | None | None | NaN | 1 | 2024-08-05 13:27:55.261966+00:00 |
| 6 | lB3IyPLQSmvt5zKv | 1 | Perform single cell analysis, integrate with C... | integrated-analysis | None | notebook | None | None | None | None | NaN | 2 | 2024-08-05 13:27:53.636178+00:00 |
| 5 | YqmbO6oMXjRj65cN | 2 | postprocess_cellranger.py | postprocess_cellranger.py | None | script | None | None | None | None | NaN | 2 | 2024-08-05 13:27:49.431280+00:00 |
| 4 | VABfOZzDfL3JFBf2 | 7.2.0 | Cell Ranger | None | None | pipeline | None | None | https://www.10xgenomics.com/support/software/c... | None | NaN | 2 | 2024-08-05 13:27:47.568616+00:00 |
| 3 | qCJPkOuZAi9q5zKv | 1 | chromium_10x_upload.py | chromium_10x_upload.py | None | script | None | None | None | None | 5.0 | 1 | 2024-08-05 13:27:45.599434+00:00 |
| 2 | T0T28btuB0PG5zKv | 1 | GWS CRIPSRa analysis | hit-identification | None | notebook | None | None | None | None | NaN | 2 | 2024-08-05 13:27:42.776119+00:00 |
| 1 | uUkmDk4oAZThcxpd | None | Upload GWS CRISPRa result | None | None | upload | None | None | None | None | NaN | 1 | 2024-08-05 13:27:39.020048+00:00 |
User
| uid | handle | name | updated_at | |
|---|---|---|---|---|
| id | ||||
| 2 | bKeW4T6E | testuser2 | Test User2 | 2024-08-05 13:27:47.562136+00:00 |
| 1 | DzTjkKse | testuser1 | Test User1 | 2024-08-05 13:27:45.107372+00:00 |
Show code cell content
!lamin login testuser1
!lamin delete --force mydata
!rm -r ./mydata
✅ logged in with email testuser1@lamin.ai (uid: DzTjkKse)
Traceback (most recent call last):
File "/opt/hostedtoolcache/Python/3.10.14/x64/bin/lamin", line 8, in <module>
sys.exit(main())
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/rich_click/rich_command.py", line 367, in __call__
return super().__call__(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 1157, in __call__
return self.main(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/rich_click/rich_command.py", line 152, in main
rv = self.invoke(ctx)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 1688, in invoke
return _process_result(sub_ctx.command.invoke(sub_ctx))
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 1434, in invoke
return ctx.invoke(self.callback, **ctx.params)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 783, in invoke
return __callback(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/lamin_cli/__main__.py", line 164, in delete
return delete(instance, force=force)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/lamindb_setup/_delete.py", line 98, in delete
n_objects = check_storage_is_empty(
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/lamindb_setup/core/upath.py", line 779, in check_storage_is_empty
raise InstanceNotEmpty(message)
lamindb_setup.core.upath.InstanceNotEmpty: Storage /home/runner/work/lamin-usecases/lamin-usecases/docs/mydata/.lamindb contains 5 objects ('_is_initialized' ignored) - delete them prior to deleting the instance
